Overview
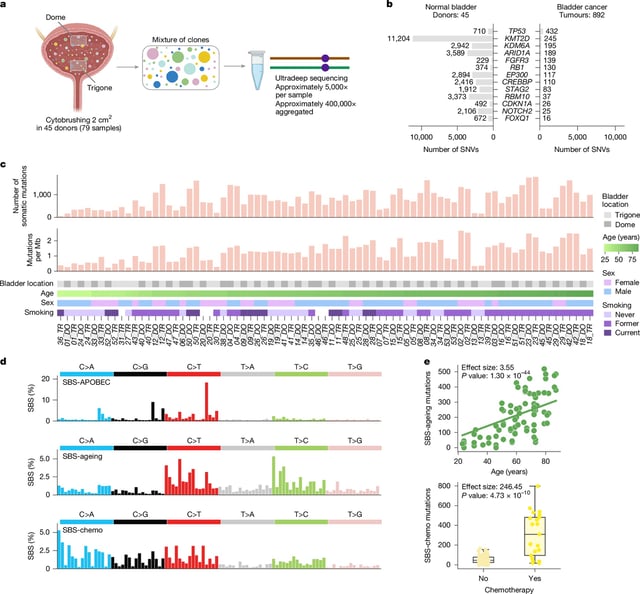
- Using targeted NanoSeq on non-invasive samples, scientists analyzed cheek swabs from 1,042 people and 371 blood samples, identifying over 340,000 mutations in cheek cells including more than 62,000 in known cancer-driver genes and 49 genes under positive selection.
- The studies documented clear mutation patterns linked to aging, tobacco use and alcohol, with smokers showing more NOTCH1 mutations and heavier clone growth and heavy drinkers displaying a distinct DNA signature.
- Most mutated cell clones in normal tissue were very small and did not keep expanding over time, indicating that common somatic mutations rarely progress toward cancer.
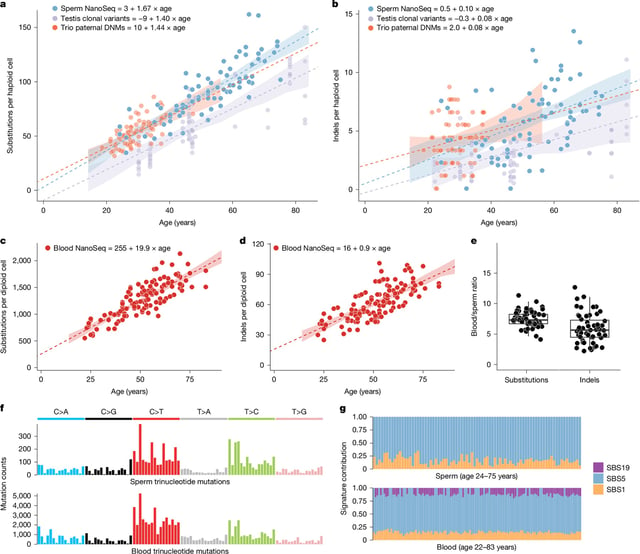
- Separate NanoSeq analysis of sperm from 81 men found an age-related rise in disease-causing mutations, from about 2% of sperm in men in their early 30s to 3–5% in middle-aged and older men, reaching around 4.5% at age 70.
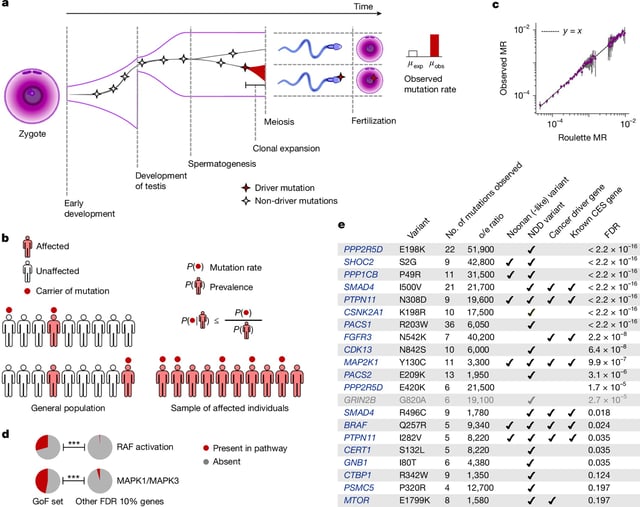
- Researchers reported extensive positive selection in the male germline, flagging roughly 40 genes in sperm and, in a complementary analysis of more than 54,000 parent–child trios, over 30 genes that gain a competitive edge, with some mutations increasing observed sperm mutation rates by roughly 500-fold.