Overview
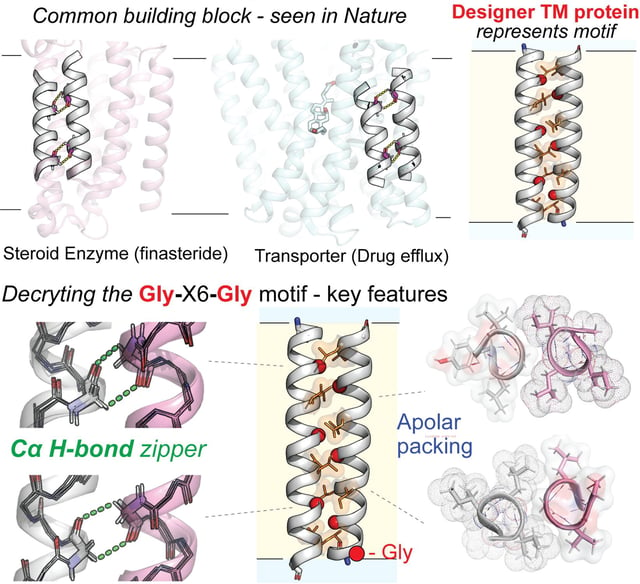
- Scripps Research reports in PNAS (Oct. 7, 2025) a computational design-and-test platform for building tractable model membrane proteins.
- The work pinpoints a conserved Gly-X6-Gly motif as a key helix–helix interaction element within lipid bilayers.
- Experiments show designer proteins folded as predicted and displayed exceptional stability, in some cases remaining intact during boiling.
- Analyses attribute the stability to repeated, individually weak hydrogen bonds that sum to create strong, motif-driven interactions.
- The team is now applying the validated platform to interpret disease-linked mutations and to design molecules that directly target membrane proteins.