Overview
- MIT engineers report prime editor variants (pPE, xPE, vPE) that cut indel byproducts dramatically, improving error rates from roughly 1-in-7 edits to as low as 1-in-543 in tested modes.
- The MIT team’s approach uses Cas9 mutations that relax nick positions to destabilize the original DNA strand, then pairs this with RNA-end stabilization to raise edit:indel ratios up to 465:1.
- HIRI researchers unveil “append editing,” a DarT2/DarG ADP-ribosylation system that tags DNA to direct repair, yielding large templated insertions in bacteria but base changes in eukaryotic cells.
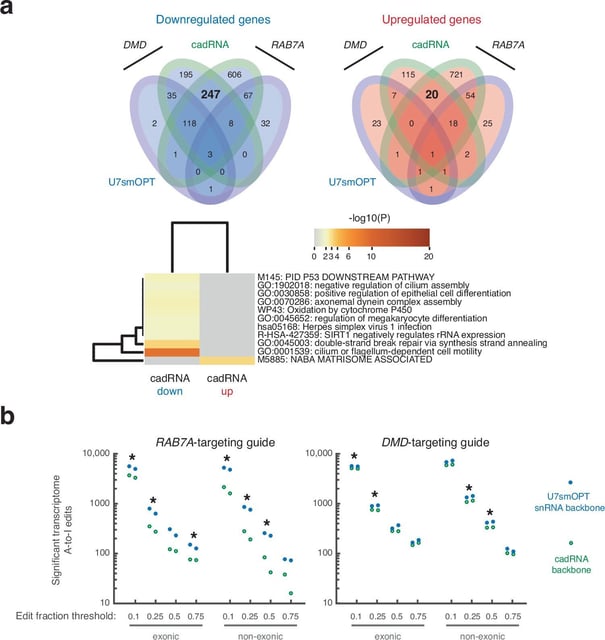
- UC San Diego and Yale introduce small nuclear RNA–guided RNA base editors that operate in the nucleus, showing stronger performance on complex transcripts, far fewer off-target edits, and functional rescue in a cystic fibrosis model.
- All three platforms are early-stage and currently validated in cells, with next steps focused on delivery, in vivo testing, and comprehensive safety assessment before any clinical use.