Overview
- The peer-reviewed study in Nature Biomedical Engineering details MARQO from a team led by Sacha Gnjatic at the Icahn School of Medicine at Mount Sinai.
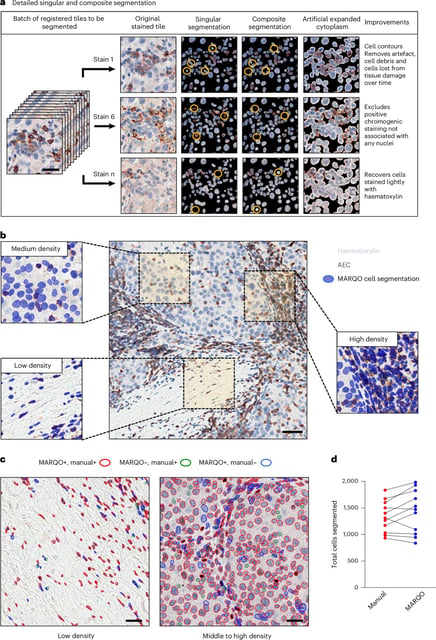
- The pipeline analyzes entire tumor slides without patching and completes runs in minutes on standard graphics cards, avoiding costly compute clusters.
- MARQO supports common immunohistochemistry and immunofluorescence staining protocols to improve comparability and reproducibility across studies.
- It automatically flags likely positive cells and records coordinates and marker intensities, with final determinations made by pathologists.
- The tool is not validated for clinical diagnostics, and the team plans UI refinements, advanced spatial and neighborhood analytics, and high‑performance computing deployments for large slide cohorts.