Overview
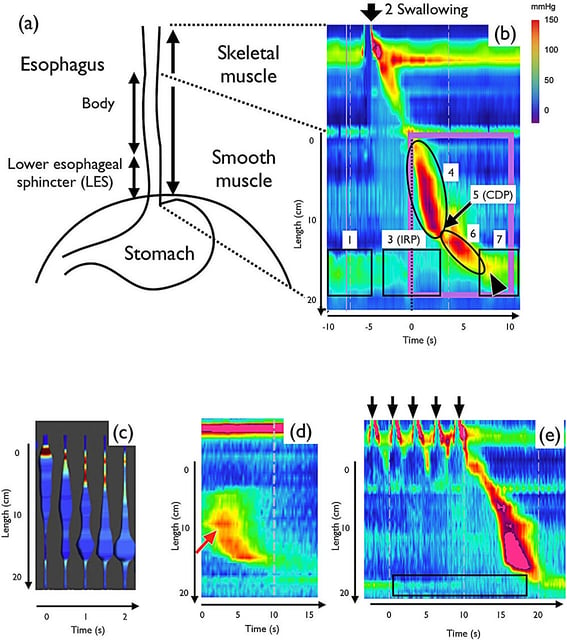
- The peer-reviewed study from Kyushu University with Josai and Hokkaido, published Aug 20 in Royal Society Open Science (DOI 10.1098/rsos.250491), uses high‑resolution manometry with simple equations to simulate esophageal peristalsis.
- The model incorporates neural signaling and lower esophageal sphincter behavior, reproducing on–off opening and deglutitive inhibition observed in clinics.
- Adjusting parameters such as nerve signal strength, firing thresholds, and muscle contraction yields outputs aligned with the Chicago Classification of motility disorders.
- Investigators say a more advanced version could help clinicians test multiple hypothesized causes per patient and support preclinical drug screening, which remains a future aim.
- The current implementation handles only liquid swallows in one dimension, with planned two‑dimensional extensions to capture complex patterns like jackhammer esophagus.