Overview
- Researchers at Harvard SEAS report a differentiable, optimization-based framework in Nature Computational Science that infers the rule sets driving morphogenesis.
- The method employs automatic differentiation to quantify how small parameter shifts in gene networks alter the final patterns formed by a cell collective.
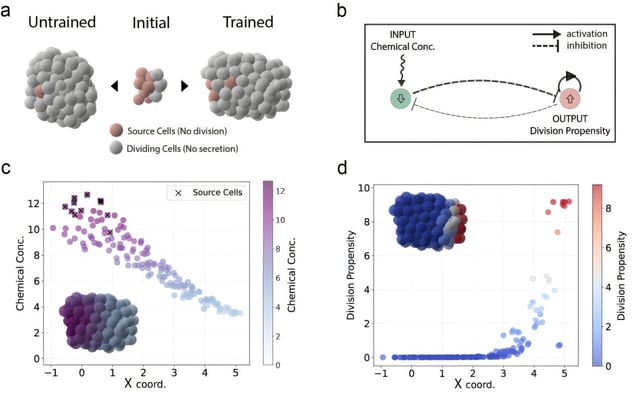
- Learned rule sets are represented as genetic networks that govern chemical signaling between cells as well as adhesion and other physical interactions.
- Proof-of-concept simulations uncovered mechanisms such as receptor-activated repression of division that, through a chemical gradient, concentrates proliferating cells at a cluster tip.
- The study, led by Ramya Deshpande and Francesco Mottes with senior author Michael Brenner, is a computational proof of concept envisioned to guide future tissue-design experiments and was supported by the Office of Naval Research and the NSF AI Institute of Dynamic Systems.