Overview
- Researchers integrated long-read sequencing of 65 diverse genomes to resolve 92% of previously missing reference sequences.
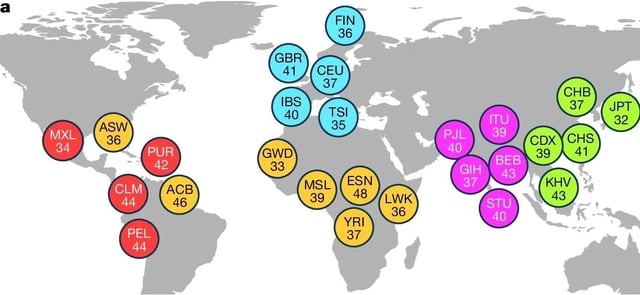
- Analysis of 1,019 individuals across 26 populations cataloged over 167,000 structural variants, with more than half of insertions and 14.5% of deletions being novel.
- High-resolution assemblies fully characterized complex, disease-linked regions including centromeres, SMN1/SMN2 genes, the major histocompatibility complex and the amylase cluster.
- The project uncovered more than 175,000 sequence-resolved structural events and doubled the known pangenome variant catalog, with all data and tools released publicly.
- Investigators emphasize the need for larger, more globally representative cohorts to capture remaining human genomic diversity and enhance clinical interpretation.