Overview
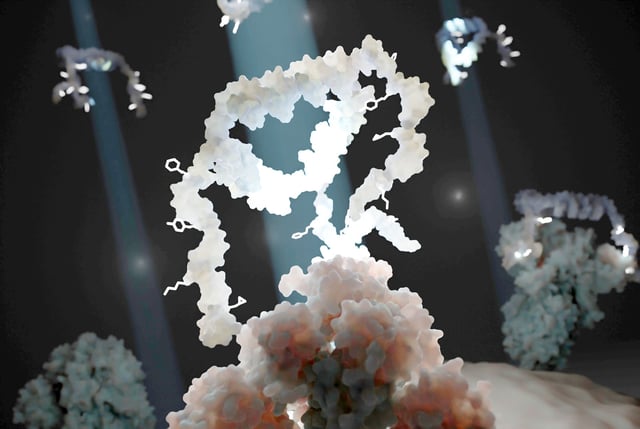
- EPFL’s Programmable Biomaterials Lab developed the MEDUSA platform to generate trimeric aptamers by organizing three binding units on protein-specific molecular scaffolds.
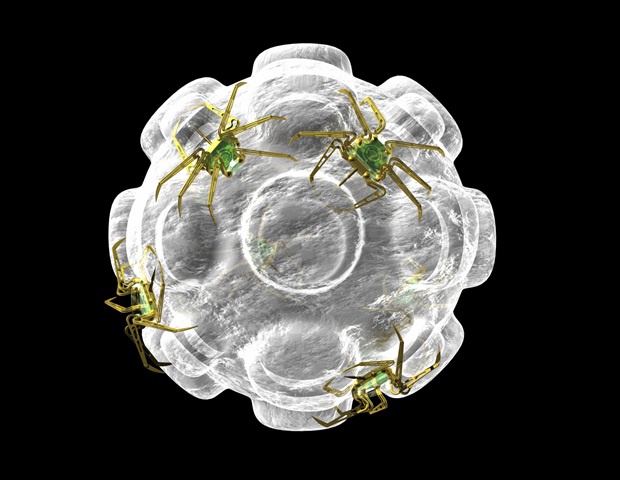
- When targeting the SARS-CoV-2 spike protein, MEDUSA-derived aptamers showed 10 to 1,000-fold stronger binding affinity than conventional monovalent binders.
- The multimeric assemblies also demonstrated markedly improved specificity, a key advantage for accurate diagnostics and antiviral therapies.
- Although scaffold design takes mere hours, the evolutionary selection process to optimize binding can span several weeks, indicating a potential bottleneck for rapid clinical use.
- Researchers plan to apply MEDUSA to pathogens with more elaborate subunit architectures like Dengue and anthrax and to harness generative AI for faster aptamer discovery.