Overview
- DynaTag outperforms existing methods such as ChIP-seq and CUT&RUN by delivering superior sensitivity and resolution in low-input and single-cell samples.
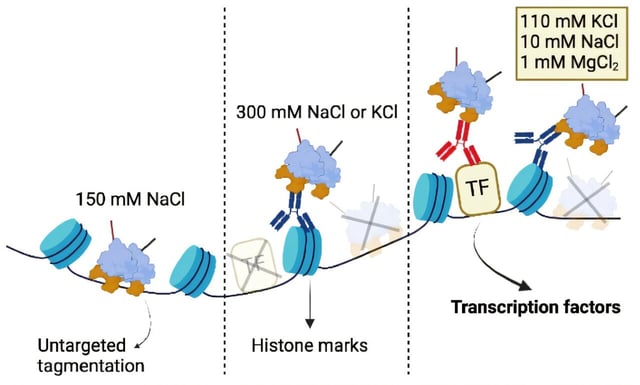
- It employs a physiological intracellular salt solution throughout all nuclei handling steps to retain specific transcription factor–DNA interactions while suppressing nonspecific binding.
- Researchers applied DynaTag to profile 15 transcription factors in mouse embryonic stem cells, revealing dynamic binding shifts of NANOG, MYC and OCT4 during cell-state transitions.
- In a patient-derived mouse model of small cell lung cancer, chemotherapy-resistant tumors showed elevated FOXA1 and MYC occupancy alongside declines in ASCL1 and POU2F3 binding.
- The method’s ability to generate high-resolution maps from scarce or clinical samples opens new avenues for studying epigenetic regulation across development, disease and treatment response.