Overview
- Nature Methods published the study on August 26, 2025, reporting superior performance versus eight existing tools.
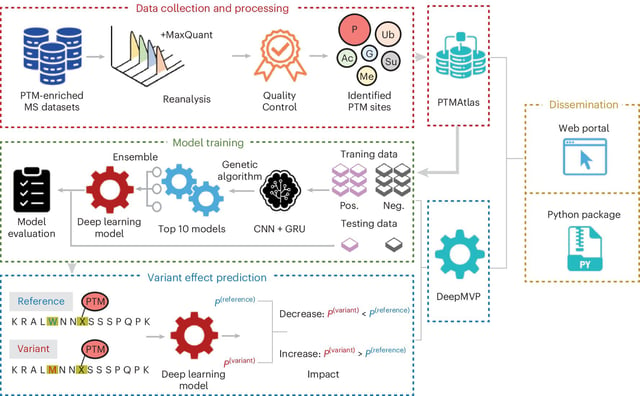
- On a curated set of 235 mutation–PTM pairs, DeepMVP predicted the affected site in 81% of cases and the direction of change in 97%.
- Predictions span six common post-translational modifications across human proteins, with support extending to viral proteins such as SARS‑CoV‑2.
- The PTMAtlas resource aggregates nearly 400,000 annotated sites across thousands of proteins to provide high-quality training and benchmarking data.
- The DeepMVP web tool and PTMAtlas are freely available to researchers at deepmvp.ptmax.org.