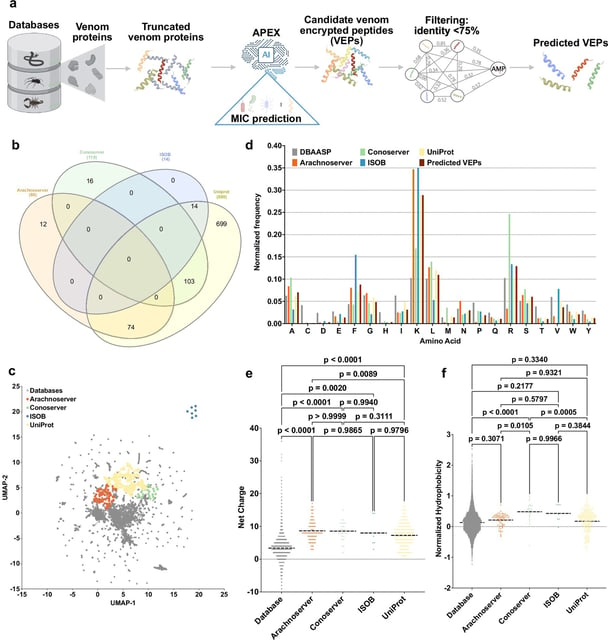
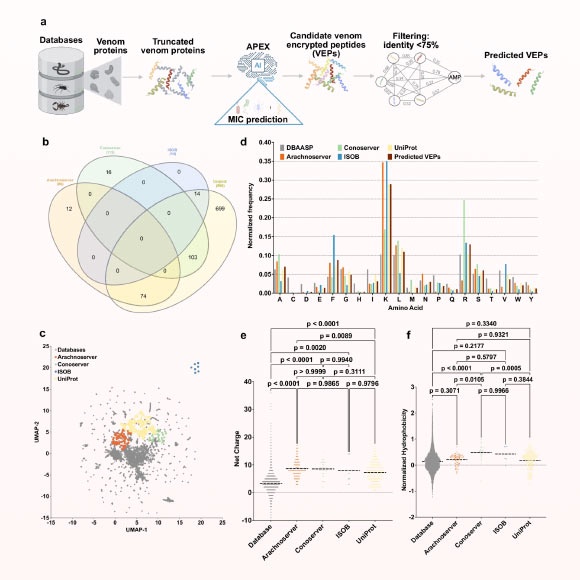
Overview
- The APEX deep-learning system scanned over 40 million venom-encrypted peptides in hours to flag 386 antibiotic candidates.
- Researchers synthesized 58 AI-selected venom peptides and found 53 that killed drug-resistant Escherichia coli and Staphylococcus aureus without harming human red blood cells.
- APEX also revealed more than 2,000 novel antibacterial motifs, expanding the structural diversity for drug development.
- Preliminary in vivo tests have commenced to assess the efficacy and toxicity of top peptide leads in animal models.
- Next steps focus on enhancing peptide stability and potency through medicinal-chemistry modifications ahead of advanced safety trials.